New molecular diagnostic methods and monitoring of microorganism genetic stability.
COVID-19 pandemic demonstrated how new whole genome sequencing technologies can help in monitoring the spread of a virus in and between populations. What is more, in almost real time we can see how virus genomes are changing when jump from one host species to another (1). Pathogen adaptation to a new host is an universal force that can be observed in baculovirus infecting insect (2) and also in coronavirus infecting different mammals (3). Broad genetic surveillance during pandemic allow us to calculate when specific mutations arose which can be further implemented in epidemiological predictions (Fig. 1).
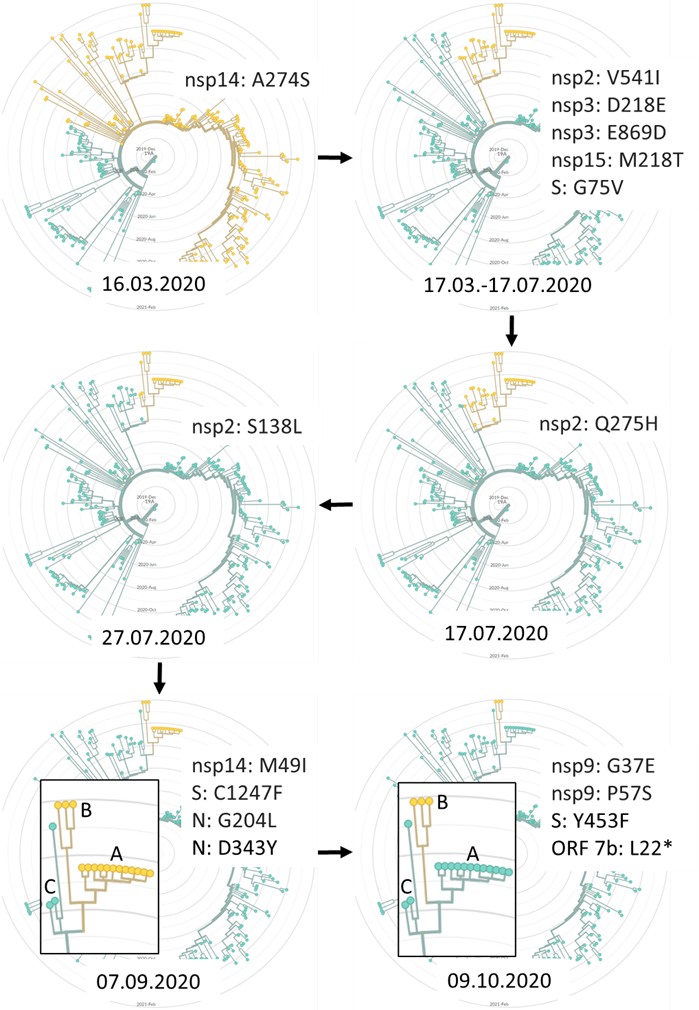
Fig. 1. Phylogenetic analysis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lineage B.1.1.279 combined with inferred time (bottom of each radial time tree) of fixing mutations (upper right of each radial time tree) in all isolates forming a group that leads to the generation of the mink variants. Yellow colour represents new variants. A: November 2020 mink isolates. B: January 2021 mink isolates and a single human isolate. C: nearest neighbours human isolates that share a common ancestor with A and BdNorway/4235/2020, Germany/NW-HHU-340/2020, and Iceland/4563/2021.
1 – Rabalski L, Kosinski M, Smura T, Aaltonen K, Kant R, Sironen T, Szewczyk B, Grzybek M. Severe Acute Respiratory Syndrome Coronavirus 2 in Farmed Mink (Neovison vison), Poland. Emerg Infect Dis. 2021 Sep;27(9):2333-2339. doi: 10.3201/eid2709.210286. PMID: 34423763; PMCID: PMC8386773.
2 – Rabalski L, Krejmer-Rabalska M, Skrzecz I, Wasag B, Szewczyk B. An alphabaculovirus isolated from dead Lymantria dispar larvae shows high genetic similarity to baculovirus previously isolated from Lymantria monacha – An example of adaptation to a new host. J Invertebr Pathol. 2016 Sep;139:56-66. doi: 10.1016/j.jip.2016.07.011. Epub 2016 Jul 22. PMID: 27451947.
3 – Rabalski L, Kosinski M, Mazur-Panasiuk N, Szewczyk B, Bienkowska-Szewczyk K, Kant R, Sironen T, Pyrc K, Grzybek M. Zoonotic spill-over of SARS-CoV-2: mink-adapted virus in humans. Clin Microbiol Infect. 2022 Mar;28(3):451.e1-451.e4. doi: 10.1016/j.cmi.2021.12.001. Epub 2021 Dec 15. PMID: 34920116; PMCID: PMC8673923.



